My current research -100,000 CT scans and only one lifetime to label them.
We’re mainlining the database! It’s the beauty of being on site with big data.
Bold italics is the journal article abstract (summary).
To say that I am hype for my current research project is an understatement. In two weeks I’m going to be presenting to the head of Radiology at my hospital (I won’t say it’s name) with a plan to publish in Nature - Scientific Data. Yeah I’m bonkers.
What we’re doing is setting up a massive project where we develop Artificial Intelligence algorithms to interpret radiological scans (X-rays, CT, MRI, Ultrasound). There’s a high barrier to entry for this kind of work. First, you need an ace IT guy to keep everything running, which we have. Then you need a steady hand at the supervisor level to help you understand the language of radiology and the game in general. Then you need at least one partner who is as hyperproductive and thirsty for a publication as you are. Check! Oh, and don’t forget that the project needs a maniacal lead who is willing to put his head on the chopping block by pushing the project like it’s the second coming.
And where there was only one set of footprints… that’s where I made a few major project decisions without telling you
As it stands now, we’re about to publish our first set of annotated data. That means we go public with our project, and our intention to become international players in radiological AI. From there, we start training our AI on our data, and posting the results.
I thought I’d lay out what the project means by talking about the abstract (the summary of the project) piece by piece.
100 Computer Tomography (CT) reports from the Emergency Department (ED) pertaining to 10 different abdominal pathologies were randomly sampled from a database spanning 20 years in one major metropolitan hospital. These reports corresponded to unique patients.
A Computer Tomography scan (CT) is a 3-D X-Ray. We use it to see inside the human body. In the ED, the CT is used a lot, because it can identify most things that are about to kill you. If you’ve ever laid in a tunnel in a hospital and got a scan - that might have been a CT. Once the scan is complete, the Radiologist looks at the images on a computer. They go through them slice by slice looking for problems. ‘Slice’, hence the ‘tomo’ in the name. You may recall another word from the same Greek derivative, atom, like a-tom, not sliceable, because atoms were thought to be indivisible. There’s a powerful counterargument to that.
Then they write a report on what they see. So the report is a description of the slices written with years, possibly decades, of experience behind it. It represents the deductive reasoning power of radiologists, who are all geniuses at my hospital. If you want to know what the CT shows, you can either train for at least 5 years in interpreting images, or you can read the report.
We went and grabbed 100 reports. But not just a random 100. We picked 10 important diseases that we’d like to study, and found 10 reports that mentioned each. We picked them all from different patients. This helped us avoid choosing useless reports.
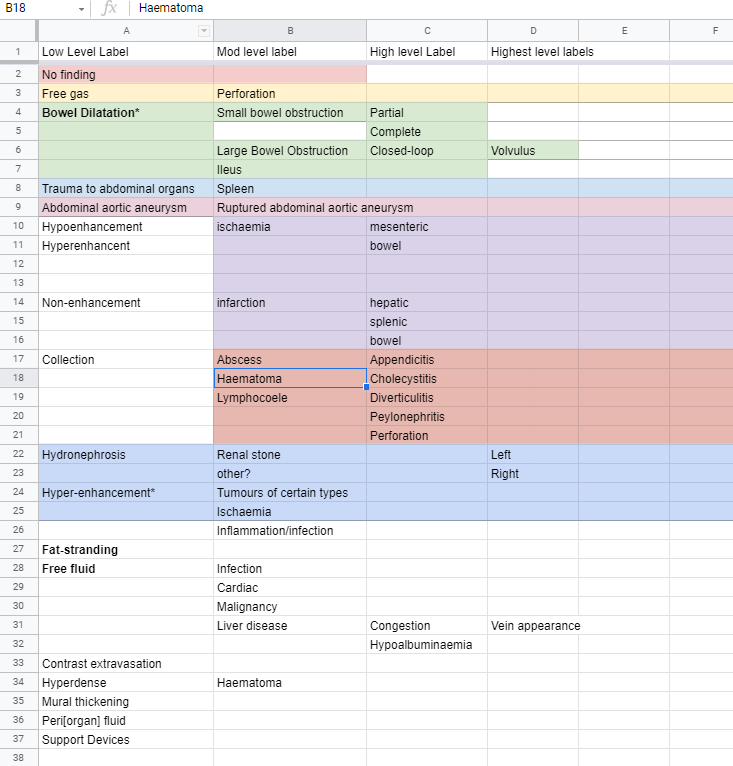
The list of diseases is here: Abscess, Appendicitis, Bowel obstruction, Cholecystitis, Diverticulitis, Laceration of Spleen, Pancreatitis, Pyelonephritis, Renal Calculus (urolithiasis), Volvulus.
Recognize many? Probably better if you don’t know.
The reports were read and annotated by a team of three medical doctors including a consultant radiologist. The reports were annotated according to findings in the following method: text spans that made mention of findings were labelled with the corresponding SNOMED Clinical Terms.
Did someone call for a doctor?
I sat down with two absolute guns and labelled each and every sentence in all 100 reports. Each report was probably 20 sentences long. Each sentence could have anywhere between 0 and 6 labels. It got pretty labour intensive. We used something called SNOMED Clinical Terms, which is like a dictionary for doctors. It helps us keep a standardized set of terms for all diseases.
So if you have a stone anywhere in your whole urinary tract (kidney, ureter, bladder, urethra) you have “Urolithiasis”. If you have a stone in the kidney, you have a “Kidney stone”. The best part is, SNOMED CT is organized so that if I say that you have a kidney stone, it understands that logically, you also must have urolithiasis! So there’s additional logical information packed into each label thanks to some clever bioinformatics work on SNOMED’s end.
We also attached each label to a phrase or two which validated that label. So if you want to check the appropriateness of that label, you can bring up the sentence quickly.
Each label was further classified by an attribute which denoted the type of mention of the finding as positive, negative, uncertain, query, past issue, or recommendation. Additionally, the laterality of the finding was denoted by another two attributes.
Ok so here’s the part where we make ourselves really useful. We don’t just record when something was mentioned. We also record how it was mentioned. You know when you meet someone and they say
“Oh, Michael, I’ve heard so much about you…”
That’s a mention.
And what you immediately start thinking is
…uh oh.
Because you want to know the mention classification. All good I hope? Bad? Ambiguous? Good in the past, but past my prime? People desperate to know more? Did you hear about my yoga or my investments? (i.e Am I on the left or the right?)
So we attached to each label for the mention a bunch of information about the disease. Especially whether it was ruled in or out, which is a major part of the radiologist’s job.
So now we have this beautiful, yet small, sample of 100 reports written by our hospital radiologist superstars and annotated by a hardcore trio of perfectionists.
The resulting dataset represents a highly accurate collection of labelled text reports which may be used to train and test natural language processing machine learning models.
The beauty of our dataset is that we’ve gone from free text, which is like an essay (like this post), to individual, standardized labels. Just the way the computer likes them. So now a computer can read through our reports and learn what we mean when we say things. It can pick up on the fact that there’s a big difference between not possibly cancer and possibly not cancer. We can even split the dataset up into sentences and phrases with their own little labels and force the computer to go through them one by one like a special quiz. This is pretty useful and there aren’t a lot of datasets like it which you can download right now.
We release this dataset with the aim that it be used by others to improve models which are designed to accurately label radiologist free text reports, as the automated labelling of large datasets is integral to the creation of quality datasets, which facilitates the training of accurate machine learning models in radiology.
If we ever want to run a deep learning algorithm over our dataset of 100000 CTs in the hospital, we need to label them all. But it took me 40 hours to label 100 reports in detail. 100000 is just not possible. So we need to get to the stage where they get labelled automatically, by an algorithm which reads the report itself. Once we have that going, we can quickly label massive datasets and start developing image-interpretation algorithms at high speed, which is where we are going with this project.
Optimistically, within a year we may have 10 interesting image-interpretation projects going concurrently, all driven by the same high quality set of initial training data. If even one of those produces something great, then it’s on like Donkey Kong.
Machine Learning. Better than video games.